Epigenetic Alterations in High Nitric Oxide-Adapted Head and Neck Squamous Cell Carcinoma Cell Lines
Author'(s): Berna Demiran1*, Burcu Yucel1, Alper Kaya1 and James A. Radosevich2
1 Department of Medical Biology, Medical School, Istanbul Medeniyet University, 34000, Istanbul, Turkey.
2 Oral Medicine and Diagnostic Sciences, College of Dentistry,University of Illinois at Chicago, Chicago, IL, 60612, USA.
*Correspondence:
Berna Demircan, PhD, Department of Medical Biology,Medical School, Istanbul Medeniyet University, 34000, Istanbul, Turkey
Received: 17 February 2019; Accepted: 04 March 2019
Citation: Berna Demiran, Burcu Yucel, Alper Kaya, et al. Epigenetic Alterations in High Nitric Oxide-Adapted Head and Neck Squamous Cell Carcinoma Cell Lines. Cancer Sci Res. 2019; 2(1); 1-5.
Abstract
Background: Head and neck squamous cell carcinoma (HNSCC) ranks as the sixth most frequent malignancy worldwide. High levels of nitric oxide (NO) has been identified in many types of human cancers including HNSCC. In this study, we investigated the effect of NO on promoter DNA methylation in cell lines of HNSCC.
Materials and Methods: The methylation status of the promoters of E-cadherin, RASSF1A and MGMT were analyzed in the parent and high nitric oxide (HNO) adapted cell lines of HNSCC using Illumina MiSequencing.
Results: We detected difference in promoter methylation levels of MGMT between the parent and HNO adapted cell lines (p<0,0001). However, treatment of the cell lines with NO did not significantly change methylation of E-cadherin and MGMT.
Conclusion: Our results suggest that NO may have diverse actions on DNA methylation. Further studies are required to clearly understand the role of NO in epigenetic alterations of HNSCC.
Keywords
Introduction
Head and neck squamous cell carcinoma (HNSCC) encompasses tumors arising from multiple sites in the head and neck regions (nasopharynx, oral cavity, oropharynx, larynx, and pharynx) and currently ranks as the sixth most frequent malignancy worldwide [1,2]. Tobacco smoking, alcohol abuse and HPV infections are important risk factors for HNSCC [3,4]. The components of tobacco have been shown to generate reactive oxygen species (ROS) and reactive nitrogen species (RNS) that may lead to lipid peroxidation and increased nitric oxide (NO) products [4,5]. It has been also suggested that stimulation of NO production by ethanol intake plays an important role in the development of head and neck cancers [4]. NO is produced by three isoforms of nitric oxide synthase (NOS); neuronal NOS (nNOS), endothelial NOS (eNOS) and inducible NOS (iNOS) [6-9]. It serves as a key signaling molecule in various physiological processes including vasodilation, central and peripheral neurotransmission, hormone secretion, inhibition of platelet aggregation, and mediation of immune response [5]. However, high levels of NO and/or expression of NOS has been identified in many types of human cancers as well as HNSCC [5,10]. NO is reported to stimulate DNA damage which results in tumor development by inducing mutations in protooncogenes and tumor suppressor genes [5]. In contrast to tumor promoting effects, NO has also been suggested to have antitumoral effects by inhibiting the cell proliferation, migration and apoptosis in various human cancers [4,10]. We have developed a laboratory model system to study the role of NO on various cancers [5,9,11-16].
Epigenetic modifications as well as genetic changes are responsible for the tumor development and progression in HNSCC [17,18]. DNA methylation is one such epigenetic event that transfers a methyl group to the C-5 position of the cytosine ring of DNA. Methylation is mainly regulated by enzymes called DNA methyltransferases (DNMTs) [1]. Aberrant DNA methylation of promoter CpG islands of some tumor suppressor genes is associated with decreased transcriptional activity in various types of cancers including HNSCC [17,18]. Many tumor suppressor genes have been indicated to be epigenetically silenced by promoter DNA hypermethylation in HNSCC and these genes are mostly responsible for regulating cell proliferation, DNA repair, apoptosis, tissue invasion and metastasis [19-24]. NO has been reported to be involved in DNA methylation as well as other epigenetic mechanisms such as histone modifications and microRNAs in normal and tumor tissues [25,26]. However, to our knowledge, NO has never been investigated regarding its possible involvement in promoter DNA methylation in HNSCC.
Given the diverse effects of NO in human cancers, understanding different actions of NO at the molecular level by epigenetic mechanisms could help obtaining NO based diagnostic or prognostic markers and lead to development of new strategies for the effective treatment of HNSCC. Thus, in this study, we aimed to investigate the effect of NO on DNA methylation in HNSCC by using a model cell line system developed in our laboratory. In the model system, cell lines of HNSCC, as well as other tumor cell lines, were adapted to high concentrations of NO over a period of months, and resultant high NO (HNO) adapted cell lines were observed to grow faster and more aggressively than each respective parent (untreated with NO) cell line [5]. By utilizing these cell lines, we analyzed and compared promoter methylation levels of tumor suppressor genes of Epithelial cadherin (E-cadherin or CDh3), Ras-Association Domain Family 1A (RASSF1A) and O-6-Methylguanine-DNA Methyltransferase (MGMT) between parent and HNO adapted HNSCC cell lines.
Materials and Methods
Cell lines and cell adaptation
All media and supplements were purchased from Invitrogen Corporation (Carlsbad, CA, USA). Both parent and HNO cell lines were maintained in MEM media supplemented with 10% fetal calf serum inactivated at 56°C for 30 min, 100 U/mL penicillin,100 μg/mL streptomycin, 2 mM L-glutamine, 2.5 μg/mL amphotericin B solution, 100 mM MEM nonessential amino acids, and 1 mM sodium pyruvate (Mediatech, Inc, Manassas, VA, USA). The five HNSCC cell lines (three originating from the tongue: SCC016, SCC040, and SCC056; one from the floor of mouth: SCC114; and one from the alveolar ridge: SCC116) were used for adaptation to the high concentrations of nitric oxide by NO donor called (Z)-1- [2-(2-aminoethyl)-N-(2-ammonioethyl) amino]diazen-1-ium-1,2- diolate (DETA-NONOate), (Sigma-Aldrich Corp., St. Louis, MO, USA). DETA-NONOate was chosen as the NO donor because of its high level of free radical donation (two moles of NO per mole of DETANONOate) and relatively long half-life (approximately 24 hrs at 37°C and pH 7.4). Each parent cell line was adapted to a high NO environment as previously described (Yarmolyuk et al., 2011). To summarize, the parent cells were treated with 50 μM DETA-NONOate, and then the concentration of donor was increased at 25 μM increments up to a point which was lethal to each parent cell line (a concentration of 600 μM). These HNO- adapted cell lines are termed SCC016-HNO, SCC040-HNO, SCC056-HNO, SCC114-HNO and SCC116-HNO, respectively.
Isolation and Sodium Bisulfite Modification of Genomic DNA Qiagen Blood and Cell Culture DNA kit (Qiagen, Inc.,Valencia, VA) was used to extract genomic DNA (gDNA) from cultured HNSCC cell lines. Isolated genomic DNA samples were undergone bisulfite deamination reaction using Qiagen EpiTect Bisulfite kit (Qiagen, CA, USA) according to manufacturer’s instructions. Briefly, 500 ng of DNA was used from each sample. Required amount of DNA in uL were mixed with 85 μL of bisulfite mix solution and 35 μL of DNA preservation buffer in 200 μL of the PCR tubes. Samples were then incubated in the thermal cycler device (BioRad, USA) for 5 hours and in changing temperatures (95°C -5 min, 60°C -25 min, 95°C -5 min, 60°C -85 min, 95°C-5 min, 60°C -175 min, respectively). Samples were transferred to the Epitect spin columns after incubation, and related buffers were added and centrifuged accordingly. Finally, bisulfite treated DNA samples were purified in 20 μL of elution buffer. One μL of bisulfite-converted gDNA solution was generally used in subsequent PCR reactions.
PCR Amplification and Sequencing of E-cadherin, RASSF1A and MGMT
PCR reactions were conducted using Qiagen HotStarTaq DNA polymerase (BioRad, USA) and supplied 1X PCR buffer supplemented with 0,1 mmol/L dNTPs, 2,5 mmol/L MgCl2, and 0,5 μmol/L each of forward and reverse primers and bisulfite- converted gDNA. The thermal cycler (BioRad, USA) was set up for initial activation step for 15 min at 95°C, and followed by 45 cycling steps of 94°C-30 sec, optimized primer-specific annealing temperature °C-30 sec, and 72°C-30 sec. PCR reaction was completed after the final elongation step applied at 72°C for10 min. Following thermocycling, PCR products were loaded 10% polyacrylamide gels and visualized using ethidium bromide. Human placental genomic DNA (Biochain Institute, Hayward, CA) was used as positive and negative control. This gDNA was either methylated in vitro with SssI methylase (NEB, Ipswich, MA) or not methylated prior to bisulfite conversion as outlined previously [27]. Samples were then prepared for sequencing by ligation of barcoded sequencing adapters using a PrepX kit implemented on an Apollo 324 robotic system (IntegenX Inc., Pleasanton, CA, USA). Barcoded adapters were NEXTflex 6nt barcodes (Bio Scientific). Sequencing was performed on an Illumina MiSeq instrument, employing V3 chemistry (600 cycles). Library preparation and pooling of samples was performed by the University of Illinois at Chicago Sequencing Core. Sequencing was performed at the W.M. Keck Center for Comparative and Functional Genomics at the University of Illinois at Urbana-Champaign (UIUC).
Wilcoxon matched-pairs signed rank test was applied to compare the methylation differences between the parent and HNO-adapted cell lines.
Results
In order to obtain a better insight into the role of NO on promoter DNA methylation in HNSCC, we first performed a comprehensive literature search regarding epigenetics of HNSCC and choose 3 tumor suppressor genes for our study. These genes (E-cadherin, RASSF1A and MGMT) have been shown to be subjected to epigenetic silencing via DNA hypermethylation in HNSCC [3,28- 30]. Primers used for PCR and sequencing analyses were designed using the online MethPrimer software (www.methprimer.org) and are listed in Table 1.
Illumina MiSeq was performed to map methylated CpG dinucleotides for the regions of 167, 188 and 198 bp (chr: 16q22.1, chr: 3p21.31, chr: 10q26.3) of E-cadherin, RASSF1A and MGMT gene promoters containing 15,16, 21 CpG sites, respectively. Raw sequence data was processed within the software package CLC genomics workbench. For each sample, from 450,303 to 827,542 clusters were acquired. Quality trimming (Q20, no ambiguous nucleotides allowed) was performed on all samples, and trimmed reads were mapped against reference ‘converted’ sequences for each gene (i.e. C positions were converted to T). Subsequently variant calling was performed using the default CLC variant caller. Variant calling tables for each sample were exported, and for each nucleotide position, the number of sequences generating each base were counted. After sequencing analysis, the Illumina reads were post-processed and aligned to the human reference regions.
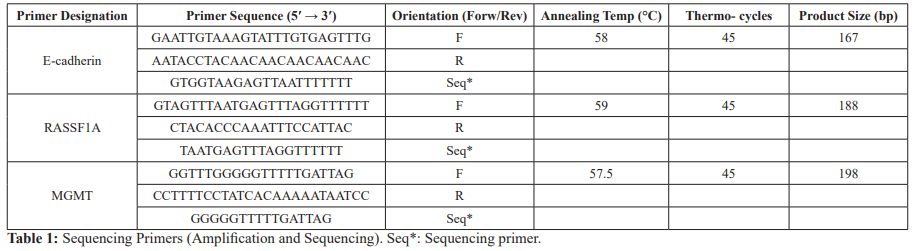
By using unmethylated and methylated human placental gDNA as a negative and positive control, respectively, we validated our results of sequencing analysis for the studied genes (Figure 1a). Promoter methylation of E-cadherin (41.7%), RASSF1A (24.6%) and MGMT (19.2%) was detected to various extents in all of the parent cell lines (SCC016, SCC040, SCC056, SCC114 and SCC116), which was in agreement with previous reports. However, according to the sequencing results, methylation levels of E-cadherin and RASSF1A did not significantly change in response to NO treatment in all of the studied cell lines. Promoter methylation status, on the other hand, of MGMT showed alterations after the treatment with NO donor in SCC016 and SCC056 cell lines. Methylation level was significantly lower in SCC056-HNO cell line than its parent cell line SCC056 (p<0,0001), whereas it was higher in SCC016-HNO cell line compared to the parent cell line SCC016 (p<0,0001) (Figure 1b). Little to no difference was detected between parent and HNO cell line pairs for the other three cell line pairs (SCC040, SCC114 and SCC116).
Discussion
NO is a highly reactive free radical molecule found in normal and malignant tissues, and have multifunctional effects in mammalian cells [5,10]. It has been reported that NO can both promote and inhibit carcinogenesis. The effects of NO in cancer cells depend on the concentration and the duration of NO exposure, and the activity and localization of NOS isoforms [31,32]. Excessive NO synthesis and NOS activity have been observed in human cancer cell lines and cells from tumor biopsies, including HNSCC [33,34]. By designing an in vitro cell line model system, we previously characterized some molecular alterations in HNO adapted cell lines of HNSCC [5,9]. Herein, we investigated the effects of high
NO levels epigenetically by utilizing same model cell line system in HNSCC. For this purpose, we detected promoter methylation levels of E-cadherin, RASSF1A and MGMT and then compared between the parent and HNO adapted cell lines (SCC016-HNO, SCC040-HNO, SCC056-HNO, SCC114-HNO, SCC116-HNO) by Illumina MiSeq platform. The methylation levels of E-cadherin, RASSF1A and MGMT in the human placental gDNA were used as negative and positive control, respectively (-SssI and + SssI) (Figure 1a). The results of placental gDNA as positive and negative controls validated our primers and sequencing analysis. According to the sequencing analysis of three tumor suppressor genes, we observed conflicting methylation levels in response to NO exposure of the cell lines.
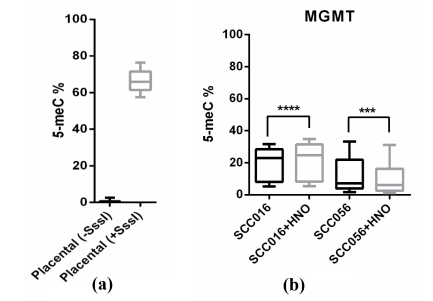
Figure 1: Methylation levels of MGMT in treated (+SsssI) and untreated (-SsssI) placental DNA (a) and in the parent/HNO-adapted cell lines of HNSCC (b).
E-cadherin is a transmembrane glycoprotein and serves as cell adhesion molecule [1]. It has been identified as a significant predictive marker of oral carcinogenesis due to its promoter methylation [28]. Our results regarding the sequence of E-cadherin promoter demonstrated DNA hypermethylation in the parent cell lines (41,7%) which were consistent with previous studies. However, treatment of these cell lines with NO did not significantly change methylation levels of E-cadherin (data not shown).
RASSF1A is a cellular tumor suppressor gene and regulates a broad range of cellular functions, including apoptosis, cell-cycle arrest, mitotic arrest, and migration. However, the expression of RASSF1A is lost in many human cancers by epigenetic silencing [35]. Meng, et al. conducted a meta-analysis and identified an association between hypermethylation of RASSF1A promoter and HNSCC [3]. Our sequencing results of RASSF1A indicated some methylation changes in the parent cell lines (24,6%), while no signicifant difference was observed between the parent and HNO- adapted cell lines.
Another tumor suppressor gene we studied was MGMT, which plays a role in the repair of DNA damage caused by alkylating agents and prevents precancerous mutations. MGMT is involved in the early and late carcinogenesis of oral cancers and is observed epigenetically silenced by promoter DNA hypermethylation in HNSCC patients [28-30]. We observed decreased methylation levels in SCC056-HNO and increased methylation levels in SCC016-HNO cell lines compared to their parent cell lines, SCC056 and SCC016, respectively (p<0,0001). Our laboratory previously performed Western Blot analysis to detect expression level of iNOS in the parent and HNO-adapted SCC016, SCC040 and SCC056 cell lines. They observed that for each cell line pair, iNOS expression was much greater in the HNO-adapted cell line than in the corresponding parent cell line [3].
Taken together, our results suggest that NO may have diverse actions on DNA methylation similar to its dual effects in tumor development and progression. Very few studies have investigated the role of NO on DNA methylation in various cancers, and these studies have demonstrated the contrasting effects of NO epigenetically. For example, Zhao, et al. reported that NO lead to a global decrease in DNMT1 and DNMT3a activity and 5-methylcytosine levels in murine squamous cell carcinoma [36]. On the other hand, Huang, et al. observed that NO induced promoter DNA methylation of tumor suppressor gene of E-cadherin in gastric cancer [37]. Another study showed that NO result in increase in DNA methylation of the tumor suppressor gene of RunX3 in gastric cancer [38]. Taking the findings of these studies and our data into consideration, it is important to evaluate various mechanisms by which NO modulate epigenetic process and thus change expression of associated genes. One such mechanism is involved in the effect of NO in the activities and expression levels of numerous epigenetic regulatory enzymes. The Ten-Eleven Translocation (TET) family are examples of these enzymes that catalyze the stepwise oxidation of 5-methylcytosine (5-mC) in DNA to 5-hydroxymethylcytosine (5-hmC). These oxidized 5-mC derivatives represent intermediates in the reversal of cytosine methylation, which is associated with active gene transciption. TET enzymes utilize iron, α-ketoglutarate and oxygen for the reaction of DNA demethylation. NO has been shown to bind to the catalytic iron and inhibit TET enzyme. Therefore, inhibition of TET activity by NO may suppress the expression of specific genes due to the accumulation of 5-mC in the promoter region [39-41].
In summary, NO appears to be an epigenetic regulatory molecule in tumor progression. However, further studies are required to clearly understand the dichotomous nature of NO epigenetically in order to facilitate its use for the diagnostic and therapeutic purposes.
Acknowledgment
This investigation was supported by the VA Merit Review Grant, USA.
References
- Wong TS, Gao W, Chan Interactions between E-cadherin and microRNA deregulation in head and neck cancers the potential interplay. Biomed Res Int. 2014; 126038.
- Lubek Head and Neck Cancer Research and Support Foundations. Oral Maxillofac Surg Clin North Am. 2018; 30: 459-469.
- Meng RW, Li YC, Chen X, et al. Aberrant Methylation of RASSF1A Closely Associated with HNSCC, a Meta-Analysis. Sci 2016; 9: 20756.
- Choudhari SK, Chaudhary M, Bagde S, et Nitric oxide and cancer a review. World J Surg Oncol. 2013; 30: 118.
- Yarmolyuk YR, Vesper BJ, Paradise WA, et Development of a model system for studying nitric oxide in tumors high nitric oxide-adapted head and neck squamous cell carcinoma cell lines. Tumor Biol. 2011; 32: 77-85.
- Peñarando J, Aranda E, Rodríguez-Ariza Immunomodulatory roles of nitric oxide in cancer tumor microenvironment says NO to antitumor immune response. Transl Res. 2019; 201: 99-108.
- Ehrenfeld P, Cordova F, Duran WN, et S-nitrosylation and its role in breast cancer angiogenesis and metastasis. Nitric Oxide. 2019; 9: 52-59.
- Salimian Rizi B, Achreja A, Nagrath D. Nitric Oxide: The Forgotten Child of Tumor Metabolism. Trends Cancer. 2017; 3: 659-672.
- Tarjan G, Haines GK, Vesper BJ, et Part II Initial molecular and cellular characterization of high nitric oxide-adapted human tongue squamous cell carcinoma cell lines. Tumor Biol. 2011; 32: 87-98.
- Shin KH, Kang MK, Park Heterogeneous nuclear ribonucleoprotein G nitric oxide andoralcarcinogenesis. NitricOxide. 2008; 9: 125-132.
- Bentz BG, Haines GK, Hanson DG, et al. Head and neck squamous cell carcinomas express a spectrum of constitutive. Biol Nitric Part 5 Portland Press. London. 1996.
- Bentz BG, Haines III GK, Hanson DG, et al. Nitric oxide synthase type 3 is Increased in Squamous Hyperplasia Dysplasia and squamous Cell Carcinoma of the Head and Neck. Ann Otol Rhinol Laryngol. 1999; 108: 781-787.
- Bentz BG, Simmons RL, Haines III GK, et al. The Yin and Yang of Nitric Oxide. Reflections on the Physiology and Pathophysiology of Head & Neck. 2000; 22: 71-83.
- Bentz BG, Chandra RK, Haines III GK, et Nitric Oxide and Apoptosis in Human Head & Neck Squamous Cell Carcinoma Development. Am J Otolaryngol. 2002; 23: 4-11.
- Chandra RK, Haines III GK, Bentz BG, et Expression of NOS3 in Reflux-Induced Lesions of the Esophagus. Otol Head Neck Surg. 2001; 24: 442-447.
- Paradise WA, Vesper BJ, GoelA, et Nitric Oxide perspectives and Emerging Studies of a Well Known Cytotoxin. Inter J Mol Sci. 2010; 11: 2715-2745.
- Németh CG, Röcken C, Siebert R, et Recurrent chromosomal and epigenetic alterations in oral squamous cell carcinoma and its putative premalignant condition oral lichen planus. PLoSOne. 2019; 14: e0215055.
- Mochizuki D, Misawa Y, Kawasaki H, et Aberrant Epigenetic Regulation in Head and Neck Cancer Due to Distinct EZh3 Overexpression and DNA Hypermethylation. Int J Mol Sci. 2018; 19: E3707.
- Strzelczyk JK, Krakowczyk L, Owczarek AJ. Aberrant DNA methylation of the p16, APC, MGMT, TIMP3 and CDh3 gene promoters in tumours and the surgical margins of patients with oral cavity J Cancer. 2018; 9: 1896-1904.
- Misawa K, Imai A, Mochizuki D, et al. Association of TET3 epigenetic inactivation with head and neck Oncotarget. 2018; 9: 24480-24493.
- Paluszczak J, Kiwerska K, Mielcarek-Kuchta D. Frequent methylation of DAB2, a Wnt pathway antagonist, in oral and oropharyngeal squamous cell carcinomas. Pathol Res Pract. 2018; 214: 314-317.
- de Freitas Cordeiro-Silva M, Stur E, Agostini LP, et Promoter hypermethylation in primary squamous cell carcinoma of the oral cavity and oropharynx a study of a Brazilian cohort. Mol Biol Rep. 2012; 39: 10111-10119.
- Kaur J, Demokan S, Tripathi SC, et Promoter hypermethylation in Indian primary oral squamous cell carcinoma. Int J Cancer. 2010; 127: 2367-2373.
- Taioli E, Ragin C, Wang XH, et al. Recurrence in oral and pharyngeal cancer is associated with quantitative MGMT promoter BMC Cancer. 2009; 9: 354.
- Nott A, Riccio Nitric oxide-mediated epigenetic mechanisms in developing neurons. Cell Cycle. 2009; 8: 725- 730.
- Hickok JR, Vasudevan D, Antholine WE, et al. Nitric oxide modifies global histone methylation by inhibiting Jumonji C domain-containing demethylases. J Biol Chem. 2013; 288: 16004-16015.
- Vo QN, Kim WJ, Cvitanovic L, et The ATM gene is a target for epigenetic silencing in locally advanced breast cancer. Oncogene. 2004; 23: 9432-9437.
- Dvojakovska S, Popovic-Monevska D, Grcev A, et Promotor hypermethylated genes Prospective diagnostic biomarkers in oral cancerogenesis. J Craniomaxillofac Surg. 2018; 46: 1737-1740.
- Onerci Celebi O, Tezel GG, Hosal AS, et al. Detection of O6- methylguanine-DNA methyltransferase gene promoter region methylation pattern using pyrosequencing and the effect of methylation pattern on survival, recurrence, and chemotherapy sensitivity in patients with laryngeal Pathol Res Pract. 2016; 212: 456-462.
- Burassakarn A, Pientong C, Sunthamala N, et al. Aberrant gene promoter methylation of E-cadherin, p16 INK4a, p14 ARF, and MGMT in Epstein-Barr virus-associated oral squamous cell Med Oncol. 2017; 34: 128.
- Fukumura D, Kashiwagi S, Jain RK. The role of nitric oxide in tumour progression, Nat 2006; 6: 521-534.
- Lala PK, Chakraborty Role of nitric oxide in carcinogenesis and tumor progression. Lancet Oncol. 2001; 2: 149-156.
- Gallo O, Masini E, Morbidelli L, et al. Role of nitric oxide in angiogenesis and tumour progression in head and neck J Natl Cancer Inst. 1998; 90: 587-596.
- Franchi A, Massi D, Santucci M, et al. Inducible nitric oxide synthase activity correlates with lymphangiogenesis and vascular endothelial growth factor-C expression in head and neck squamous cell J Pathol. 2006; 208: 439-445.
- Dammann RH, Richter AM, Jiménez AP, et al. Impact of Natural Compounds on DNA Methylation Levels of the Tumor Suppressor Gene RASSF1A in Cancer. Int J Mol Sci. 2017; 18: 2160.
- Zhao H, Ning S, Scicinski J, et al. Epigenetic effects of RRx- 001 a possible unifying mechanism of anticancer activity. 2015; 6: 43172-43181.
- Huang FY, Chan AO, Rashid A, et al. Helicobacter pylori induces promoter methylation of E-cadherin via interleukin- 1β activation of nitric oxide production in gastric cancer Cancer. 2012; 118: 4969-4680.
- Katayama Y, Takahashi M, Kuwayama Helicobacter pylori causes runx3 gene methylation and its loss of expression in gastric epithelial cells, which is mediated by nitric oxide produced by macrophages. Biochem Biophys Res Commun. 2009; 388: 496-500.
- Branco MR, Ficz G, Reik Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2011; 13: 7-13.
- Kohli RM, Zhang TET enzymes TDG and the dynamics of DNA demethylation. Nature. 2013; 502: 472-479.
- Vasudevan D, Bovee RC, Thomas DD. Nitric oxide the new architect of epigenetic Nitric Oxide. 2016; 30: 54- 62.